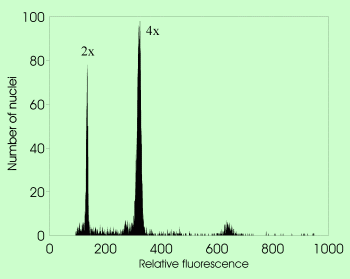
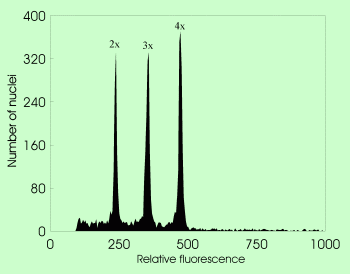
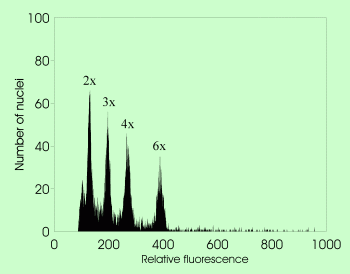
: DNA content of the unreplicated haploid nucleus (irrespective of the ploidy level)
Homoploid genome size (Cx-value): DNA content of the monoploid chromosome set (in polyploids averaged)
Holoploid and homoploid genome sizes equal in diploid species, however, the former exceeds the latter in polyploid species. Both values can be expressed either in DNA picograms or megabase pairs (1 pg = 978 Mbp).
Genome size data are useful for:
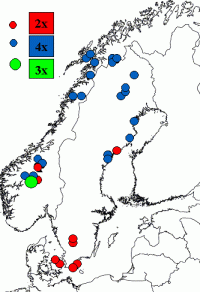
- identification of species with the same number of chromosomes
(species-specific genome size occurs in several plant alliances)
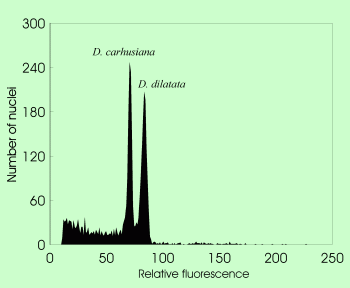 | Discrimination between species with the same number of chromosomes but different genome size in Dryopteris dilatata alliance |
- detection of hybrids from homoploid crosses
(providing the putative parents differ enough in genome size, FCM allows reliable detection of their homoploid crosses)
- assessment of genomic constitution in allopolyploid taxa or identification of putative parents
(in some hybridogenous groups, identification of their progenitors is possible)
- wide-range utilization of genome size data
(genome size allows to predict various phenotypic characters, or phenological and ecological behaviour)
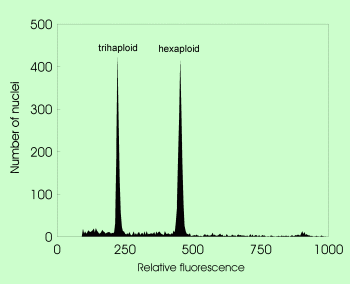
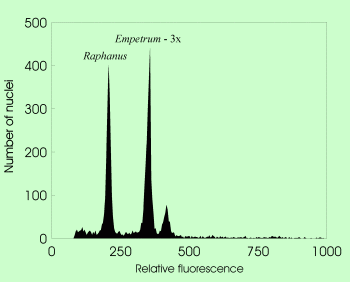 | Genome size determination in triploid Empetrum using Raphanus sativus as internal standard |
- base composition
(knowledge of AT/GC base proportion may provide further, more profound, insight into genome organization)
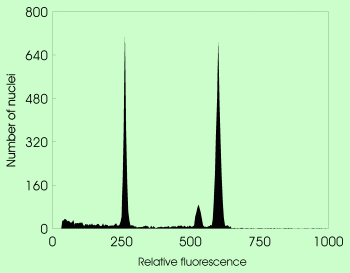 Simultaneous analysis of tetraploid Hieracium cymosum and Zea mays as internal standard stained with DAPI (fluorochrome with preferential binding to A-T base pairs) | 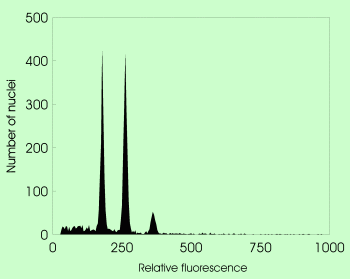 The same species pair stained with propidium iodide (intercalating fluorochrome with no base preference). Note marked difference between peak positions in both analyses. |
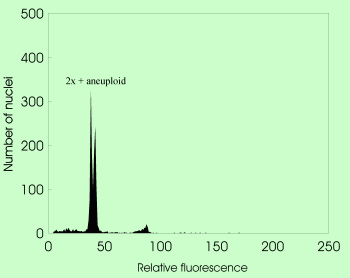
- agmatoploidy
(non-lethal chromosome fragmentation owing to the presence of diffuse centromere leading to the increase of chromosome number under constant holoploid genome size)
A revolutionary advance for efficient reproduction pathway screening, based on different proportional DNA contents of embryo and endosperm in mature seeds. Considering the type of male and female gametes (reduced vs. unreduced), the embryo origin (zygotic vs. parthenogenetic), and the endosperm origin (pseudogamous vs. autonomous route), ten different pathways of seed formation can be reconstructed.
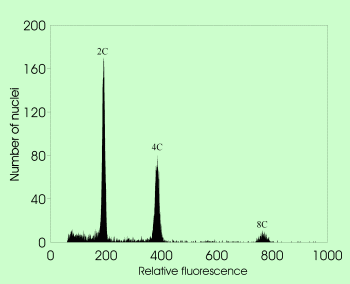
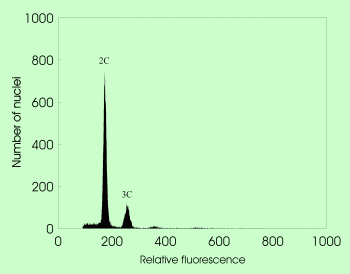 Flow cytometric acquisition of mature seeds in sexual Taraxacum with 2C embryo and 3C endosperm nuclei | 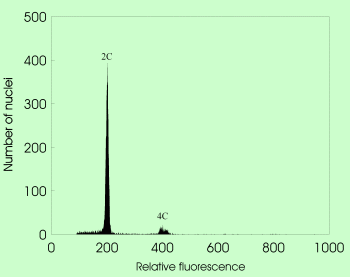 Flow cytometric acquisition of mature seeds in apomictic Taraxacum with 2C embryo and 4C endosperm nuclei |